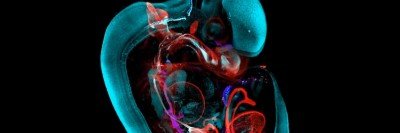
A Developing Mouse Embryo
To study a complex ecosystem, researchers often deploy an armada of high-tech tools.
On the African savanna, for example, drones track migrating elephants. Infrared sensors spy on leopards as they prowl at night. And DNA tests reveal the unique genetic fingerprints of plant and animal species. Analyzing all this data requires enormous computing power.
And so it is for MSK researchers, as they investigate the hugely complex ecosystem of cancer in the human body. To advance their understanding, MSK scientists are pushing the frontier of what technology can see, on a scale that stretches from the atom to the entire human organism.
What they are discovering is already changing how MSK — and the world — treat cancer.
Guided Tour
“This is an image that I probably couldn’t have shown you a year ago,” Anna-Katerina “Kat” Hadjantonakis tells an audience on a February afternoon. She is the Chair of Developmental
Biology at the Sloan Kettering Institute (SKI) and Alfred P. Sloan Chair and is leading a guided tour through a 3D image of a mouse embryo.
“The mouse tissue is dense and opaque,” says Dr. Hadjantonakis. “But the resolution of the imaging is so high that we can zoom in to see any individual cell in the context of the tissue surrounding it.”
That means, she explains, that “we can study cells not just one at a time, but by the tens of
thousands. We can see all the cells that make up an organ at the same time.”
She continues that “the tissues of our body are infiltrated with blood vessels and lymphatic vessels and the immune system, to name just a few. They are very complex. Now we can look at all these different features at once — and over time.”

Movie Magic
This breakthrough is achieved by microscopes that look nothing like the device many people peered through in high school.
The 3D mouse image was produced by the Bruker Luxendo MuVi SPIM light-sheet microscope. It works by taking thousands of scans using a sheet of laser light at different depths, and from different positions, and then digitally compiling them into a single 3D image. Dr. Hadjantonakis compares the resulting image to a movie — while older technologies produced snapshots.
“Imagine a 90-minute movie on Netflix,” she says. “If you were only given one snapshot of the movie every five minutes, it would be very difficult to piece together the story.” With the new technologies, however, “we can look at all the actors together and see how they interact. And we can look at them over time to see how the story unfolds. That means we can make new insights and raise new questions.”
As a developmental biologist, one of the big questions that fascinates Dr. Hadjantonakis is the process by which normal cells grow and work together to build organs — and “how those processes are disrupted or hijacked in diseases like cancer.”
She points out that “tumors are not entirely composed of cancer cells. They are actually an integration of tumor and normal cells. It’s really important that we understand how both normal and cancer cells arise — and how they interact in a tumor.”
The goal, she says, “is to understand what’s normal, what’s going wrong in disease, and how we can intervene.” Long-term, the insights gained into how single cells develop into tissues may provide the ability to “repair, replace, or regenerate an organ damaged by cancer, so we can restore patients to their normal lives.”
How Immune Cells Sink Their Fangs
Morgan Huse, a member of the Immunology Program at SKI, wields cutting-edge imaging tools to study another key component of the cancer ecosystem — the interaction between tumors and the immune system.
One of his focuses is the immune system soldier called a T cell, which attacks harmful pathogens, including cancer. Dr. Huse explains that “the notion had been that a T cell stuck itself onto the target cell and kind of vomited toxic molecules onto the surface of the target cell, hopefully killing it.”
However, that idea was challenged when Dr. Huse and his team, armed with super-resolution microscopes like the Zeiss Elyra, looked more closely at T cells in the act of killing. They found that T cells actually exert mechanical force against target cells, squeezing them “like a stress ball,” according to Dr. Huse. The T cell inserts its toxin very precisely into the target cell using “a highly evolved physical delivery system” not unlike how a snake uses its fangs to inject venom.
The result, says Dr. Huse, “is that the T cell maximizes its killing power while using less toxin and lowering the chance that the toxin will leak out and hurt an innocent bystander cell.”

Finding Vulnerabilities
To capture these “very small and very fast interactions,” the Zeiss Elyra microscope casts light into a pattern like a crisscross lattice, then rapidly moves the pattern and combines all the resulting images to extract a higher-resolution image.
Having revealed a critical mechanical dimension to T cell killing, Dr. Huse’s research is now investigating the implications. “It turns out that metastatic cancer cells need to physically stiffen in order to colonize new organs. In collaboration with Joan Massagué’s group, we recently showed that this stiffening makes the cancer cells more vulnerable to T cells.”
This new type of biomechanical vulnerability could perhaps be given a boost by medicine in the never-ending arms race between the immune system and cancer.
T Cell Attack
An Intersection of Revolutions
This new world of research relies on one of the fastest-growing fields in biomedical science: computational biology and related disciplines. Dr. Hadjantonakis points out that it takes “very sophisticated computational methods to analyze all the data we’re generating — a computer may extract patterns that I may not see.”

That’s where computational biologists come in, like Christina Leslie, a member of the SKI Computational and Systems Biology Program. Trained as a mathematician, she helped develop crucial artificial intelligence and machine learning methodologies that make sense of the enormous data sets generated by research into cancer genetics and the human genome — the blueprint of genetic instructions.
Today, Dr. Leslie’s field will be just as important in unlocking the secrets hidden in the mountain of data generated from the focus on the cancer ecosystem. She says: “I think we’re at the intersection of multiple revolutions. There’s a revolution in single cell genomics. There’s a revolution in genome editing, which allows us to study the consequences of genetic changes. There’s a revolution in data science. And it’s all moving really fast.”
Investment for the Future
Staying ahead of the revolution is the purpose of the Research Technology Transformation (RTT) initiative at SKI, which relies on generous support from donors, including the Friedman Family Foundation and Cycle for Survival®. The RTT seeks to get the best new technology into researchers’ hands quickly and share resources when possible. That provides a competitive edge that is essential to recruit and train the best and brightest of tomorrow.
Dr. Hadjantonakis says: “I don’t think we would have been able to bring in these instruments that will allow us to make these scientific advances without the RTT. It puts us five years ahead of the curve of other institutions.”
Several high-profile projects in the pipeline include:
- Triple Quadrupole Mass Spectrometry, led by Derek Tan (Eugene W. Kettering Chair), which will cut sample processing time from 10 minutes to as little as six seconds.
- Expanding the Single Cell Analytics Innovation Lab (SAIL), led by Dana Pe’er (Alan and Sandra Gerry Chair) and Ronan Chaligné.
- Organoid Innovation Lab, which will create disease models using patients’ own cancer cells, under the direction of Scott Lowe (Geoffrey Beene Chair), Lorenz Studer, and Charles Sawyers (Marie-Josée Kravis and Henry R. Kravis Chair in Human Oncology and Pathogenesis).
Together, all these resources will search for vulnerabilities in the hugely complex ecosystem of cancer in the human body, another major step forward in the effort to ensure that cancer is no longer the king of the jungle.