Our chromosomes are much longer than the space in which they are packaged in the cell. Chromosome compaction is on the order of several thousand fold, yet these chromosomes have to be unraveled every cell cycle to be replicated accurately and the daughter chromosomes must be topologically unlinked to allow their separation and segregation into the daughter cells. Management of chromosome shape and topology in bacteria falls to three groups of proteins: the DNA topoisomerases, the structural maintenance of chromosome proteins (SMC), and the small double-strand DNA-binding nucleoid-associated proteins. Our interest in these processes stems in large part from our observation that the major cellular chromosomal decatenase, Topoisomerase IV, and the cellular condensin, MukB (Fig. 5), interacted physically and that this interaction stimulated the activity of Topo IV.
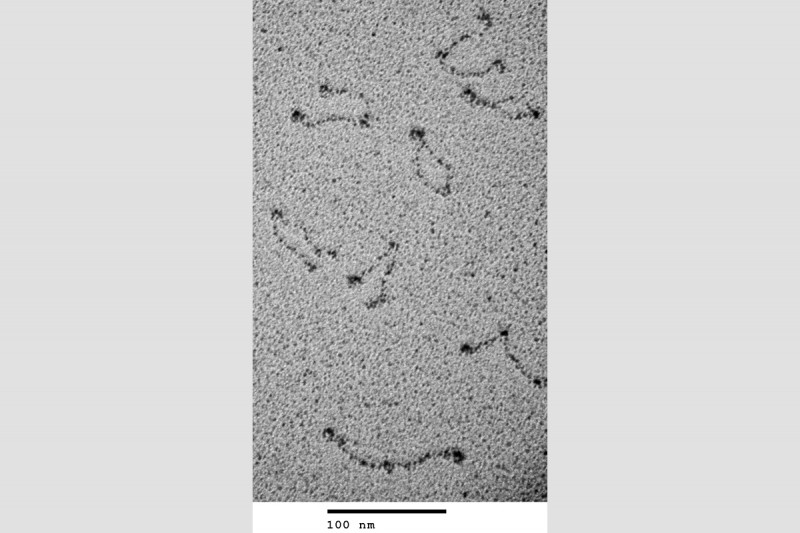
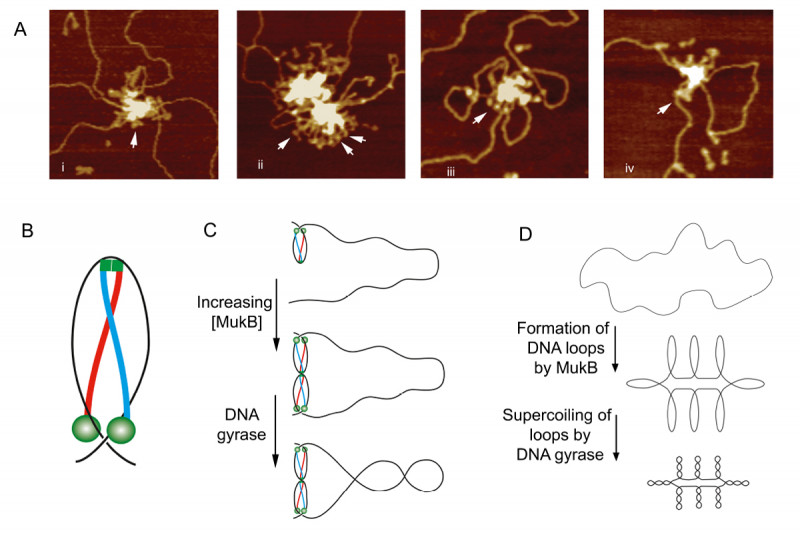
We developed gel electrophoresis-based assays that could be used to easily score DNA condensation. In these assays, a large nicked DNA plasmid when bound by MukB migrated faster in the gel than the plasmid alone. Higher concentrations of MukB caused the plasmid-DNA complex to move more slowly than the DNA alone, as expected. We showed that the faster-moving complex was a result of MukB stabilizing negative supercoils in the topologically unconstrained plasmid and that the slower-moving protein-DNA complex was a result of MukB stabilizing large relaxed loops in the DNA. We demonstrated that loop stabilization was the result of hinge-hinge interactions between MukB dimers. This led us to propose a model (Fig. 6) for MukB-mediated DNA condensation that was distinct from the prevailing view arguing that SMC proteins condense DNA by topologically trapping the DNA in the tripartite complex formed by the SMC protein and its accessory proteins. We showed using two different assays that this latter model was not operative for MukB.
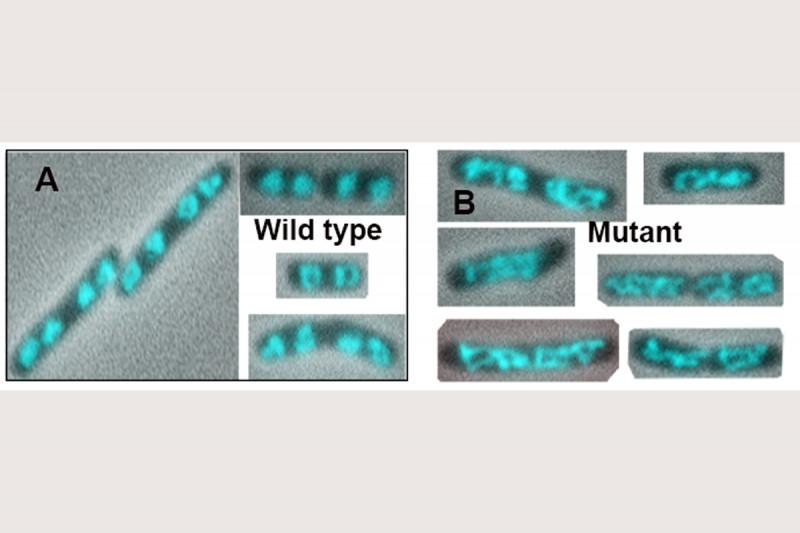
The addition of Topo IV to these MukB DNA condensation assays resulted in increased DNA condensation. This hyper-condensation of the DNA required the MukB-Topo IV interaction, but not the catalytic activity of Topo IV, suggesting that the combination of Topo IV and MukB might be serving as a scaffold for DNA compaction. This hypothesis is supported by the observation that replacing the wild-type mukB allele in E. coli with an allele (mukBtriple) encoding a mutant MukB that cannot interact with Topo IV severely disrupts the disposition of the chromosomes in the cell (Fig. 7).
Remarkably, we have recently determined that in the hyper-condensed state Topo IV is also trapping and stabilizing negative supercoils. This observation is counter to current dogma with respect to Topo IV mechanism that asserts, based on structural data, that the enzyme traps a positive supercoil. We are currently trying to reconcile our observations with Topo IV mechanism. We are also currently examining the nature of the MukB-MukE-MukF-Topo IV complex on DNA and distal effects of the interaction between the ParC subunit of Topo IV and the MukB hinge region that affect MukF binding to the neck of the MukB coiled-coil domain to modulate the ATPase activity of MukB, which is essential for DNA condensation.
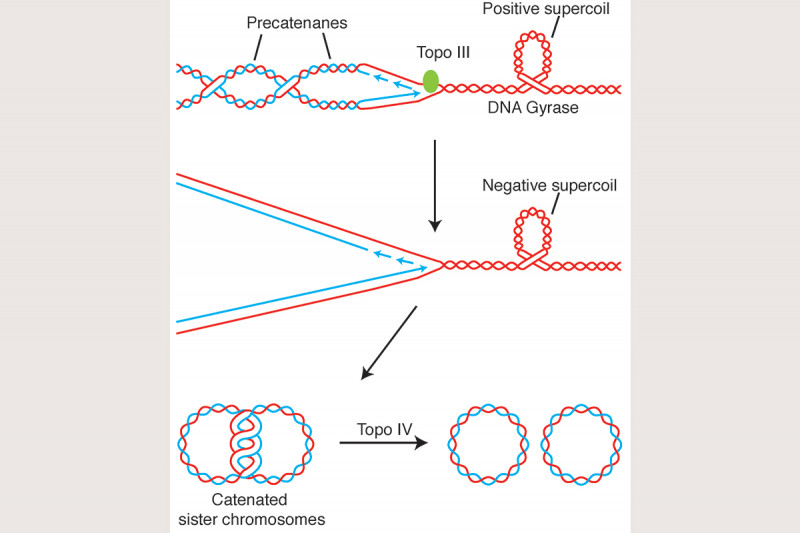
We have also argued that unlinking of the sister chromosomes during DNA replication is aided by removal of pre-catenanes that are forming directly at or just behind the replication fork (Fig. 8) and that in E. coli precatenane unlinking can be attributed, at least partially, to the action of Topoisomerase III. In support of this hypothesis, we have shown that Topo III is localized in the cell to the vicinity of the replisome and that inactivation of Topo III in vivo leads to disrupted nucleoid separation. This argues strongly that complete chromosome unlinking during the cell cycle is a combination of the action of Type I and Type II topoisomerases.